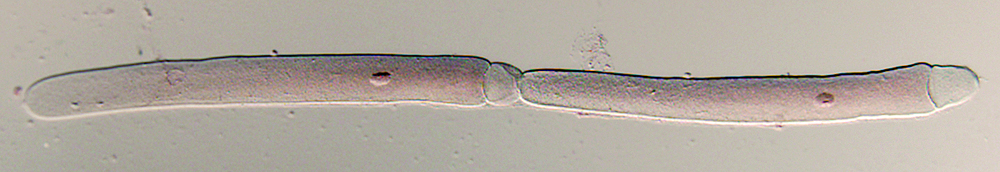
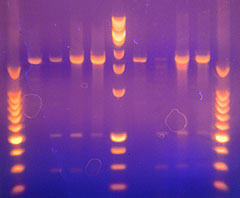
Speciation and molecular phylogeny of gregarines
Our laboratory has proven expertise in gregarine molecular phylogeny at the supra-specific level, but most of the questions we are currently addressing center on speciation and species-isolating mechanisms in gregarines. This work will be strengthened and informed by a robust molecular phylogeny at the species level, in particular, assorting species of Blabericola and Protomagalhaensia. Such a phylogeny has required the development of primer sets that allow access to additional genes for molecular phylogenetic analysis, in particular, genes with sufficiently variable regions to robusting discriminate gregarine species within a single genus. We have developed primers and techniques using the existing EST data base for Gregarina niphandrodes and a new whole-genome lumina sequence data base forBlabericola migrator. Using these primers and genes, we are currently acquiring and assembling molecular data for the 5 known species of Blabericola and the 4 known species of Protomagalhaensia. As our laboratory describes new species in these genera (6 new species descriptions are in progress), we will add their molecular data to the analysis. Given the close relationship of these gregarines, this system should provide sufficient phyogenetic reolution to resolve speciation patterns in the group and the underlying factors influencing speciation in the group.
Deb Clopton is the lead researcher on this project.